Main content
Top content
- P1 - Roland Brandt
- P4 - Michael Hensel
- P5 - Jürgen Klingauf
- P7 - Achim Paululat
- P8 - Jacob Piehler
- P11 - Christian Ungermann
- P13 - Sabine Zachgo
- P14 - Joost Holthuis
- P16 - Karin Busch
- P17 - Daniel Kümmel
- P18 - Roland Wedlich-Söldner
- P19 - Christian Kost
- P20 - Florian Fröhlich
- P21 – Heiko Harten
- P22 – Milos Galic
- P24 - Ayelen Gonzalez Montoro
- P25 - Caroline Barisch
- P26 - Katia Cosentino
- P27 - Arne Möller
- INF - Michael Hensel
- Z - Karin Busch, Michael Hensel, Jürgen Klingauf, Stefan Kunis, Jacob Piehler
- IRTG - Achim Paululat, Joost Holthuis, Roland Brandt, Christian Kost, Sabine Zachgo
- Former Projects (2011 - 2018)
- P2 - Guy Hanke
- P3 - Jürgen Heinisch
- P6 - Hans Merzendorfer
- P9 - Renate Scheibe
- P10 - Heinz-Jürgen Steinhoff, Johann Klare
- P12 - Helmut Wieczorek
- P15 (old) - Sabine Hunke
- P15 (new) - Michael Hensel, Katharina Miskiewicz
Project P1 - Roland Brandt
Regulation of tau localization and expression by neuronal RNA granules as dynamic microcompartments
Neurons utilize different mechanisms to ensure a compartment-specific protein distribution, which is vital for the proper function of highly polarized cells such as nerve cells. An example is the microtubule-associated protein (MAP) tau, which is enriched in the axon in healthy neurons and distributes to the somatodendritic compartment during a class of neurodegenerative diseases collectively called tauopathies. Mechanisms which lead to an enrichment of tau in the axon are thought to involve transport and compartment-specific binding of the protein but also mechanisms, which operate on the level of the mRNA such as regulation of translation and transport of tau mRNA in RNA granules may contribute. RNA granules can be considered as dynamic cellular microcompartments that contain a subset of different mRNA species and various mRNA-binding proteins. The project will focus on the mechanisms by which the three tau-mRNA binding proteins HuD, IMP-1 and G3BP-1 affect tau localization and expression in neurons. The dynamic interaction of the three RNA-binding proteins in RNA granules will be analyzed in detail. Subsequently, the effect of external stimuli that are relevant for neuronal functioning as well as the influence of conditions being involved in degenerative processes will be determined. Furthermore, additional components of tau-mRNA containing granules will be identified and functionally characterized. Long-term goal of the project will be to visualize and manipulate tau expression also in a systemic context (organotypic slice preparations, mouse brain) by affecting the function of tau mRNA-containing neuronal granules as a potential drug target for the treatment of neurodegenerative diseases.
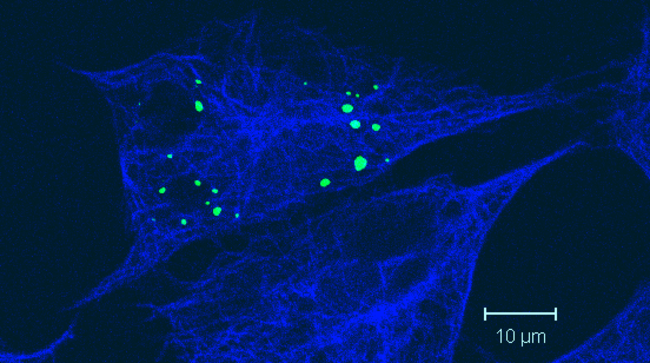
Distribution of RNA granules and microtubules in cultured cells.
Project P4 - Michael Hensel
Cooperation of two protein secretion systems during the interaction of Salmonella enterica with host cells
Salmonella enterica is an invasive, facultative intracellular gastrointestinal pathogen. We are interested to understand the molecular mechanisms deployed by Salmonella for invasion of polarized epithelial cells. Salmonella uses the SPI4-T1SS secreted giant adhesin SiiE for temporal binding to the apical side of host cell. This intimate contact allows translocation of effector proteins by the SPI1-T3SS. Activity of the effector proteins result in effacement of the brush border, induction of massive membrane ruffles and formation of novel reticular F-actin structures in a highly dynamic manner. We propose an adhesion/invasion microcompartment at the host-pathogen interface with SiiE-mediated membrane contact sites, functional effector translocation and recruitment of the host cell actin organizing machinery. The project will focus on the functional and structural analyses of the interaction microcompartment. To address these issues, we work on four aspects: a) Ultrastructural analysis of the interaction microcompartment. b) Mechanism of host cell binding by the giant adhesin SiiE. c) Characterization of bacterial and host cell proteins involved in the formation of the adhesion/invasion microcompartment. d) Mechanisms of host cell actin remodeling. The results obtained for these subprojects should reveal structural and functional details of the dynamic adhesion/invasion microcompartment formed during Salmonella invasion of host cells.
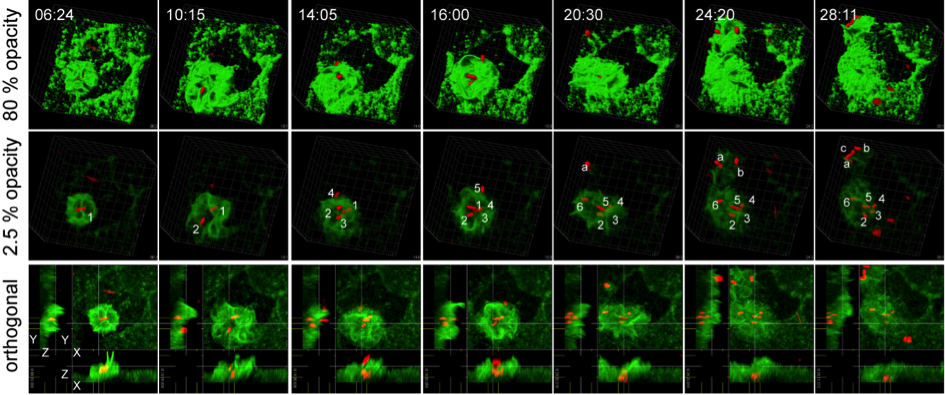
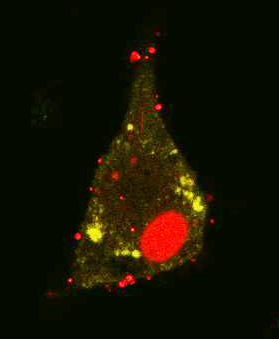
Live cell imaging of cooperative Salmonella invasion. MDCK cells permanently transfected for expression of LifeAct-GFP (green) were grown as polarized monolayers and infected with Salmonella expressing mCherry (red). The time-lapse series was recorded using a spinning disk microscope Massive membrane ruffles are initiated after contact to a single bacterium and subsequently, further bacteria cluster at the side of invasion. Numbers indicate individual bacteria. Time stamp: min:sec.
Project P5 - Jürgen Klingauf
Tethering of secretory vesicles studied by high and super-resolution microscopy
Exocytosis of secretory vesicles depends on the consecutive action of tethering factors and SNARE proteins for fusion. In mammalian cells the SM protein Munc18-1, which interacts with the SNARE protein syntaxin, is an essential component of the tethering machinery. We will use high and super-resolution fluorescence microscopy like total internal reflection fluorescence microscopy (TIRFM) and fluorescence photo-activation localization microscopy (FPALM) to visualize and track the spatio-temporal dynamics of tethering and SNARE proteins in live and fixed cells with single molecule resolution.
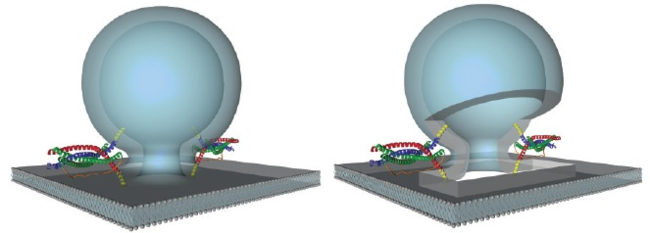
An illustration of SV fusion driven by two SNARE complexes during neuroexocytosis. Assembly of two SNARE complexes by coil-coiling of the vesicle SNARE protein syb2 (blue) and the two presynaptic membrane SNARE proteins syntaxin-1A (red) and SNAP-25 (green) drives fusion. Each SNARE complex consists of four α-helices aligned in parallel, with syb2 and syntaxin-1A contributing one helix and SNAP-25 contributing two helices. The transmembrane regions of the SNARE proteins are depicted in yellow (better seen in the version with a cut-open fusion pore to the right).
Project P7 - Achim Paululat
Role of metalloproteases in extracellular microcompartment formation and function in Drosophila heart and muscles cells
The extracellular matrix (ECM) and the proteins that constitute it are essential for providing structural support and participate in cell adhesion, migration and tissue morphogenesis. Due to its diverse physiological functions, the ECM has to be more than a homogeneous mass of proteins and carbohydrates. Within the meshwork of its structural components, the ECM is spatially patterned and thereby provides locally restricted reaction environments and structural microcompartments. The assembly, regionalization and remodeling of the ECM is a highly dynamic process, and members of the large family of metalloproteases are considered to be mandatory for functionalization of the ECM. In this project, we aim to analyze the physiological and developmental functions of two metalloproteases acting at distinct surface regions of heart and muscle cells in the genetically accessible in vivo model system Drosophila.

Project P8 - Jacob Piehler
Molecular and cellular regulation of JAK/STAT signaling
Functional selectivity is a prominent feature of cytokine signaling, which has been related to affinity and kinetics of ligand-receptor interactions. To unravel how ligand binding governs specificity of signal activation we have developed single molecule imaging technique to probe assembly and dynamics of cytokine receptors in the plasma membrane of living cells. These studies revealed ligand-induced receptor dimerization, which promotes intrinsic interactions between the associated Janus kinases (JAKs). Based on a detailed energy landscape obtained from quantitative dimerization assays, we have generated a structural model of a transmembrane cytokine receptor signaling complex. We now aim to validate our mechanistic model and to correlate receptor assembly with downstream signal activation. Using different cytokine receptor model systems we will investigate structural organization and dynamics of JAKs. We will explore the dynamics of receptor dimerization at the plasma membrane and in endosomes by single molecule FRET and we will identify the molecular and cellular determinants of STAT activation by chimeric and synthetic receptors.
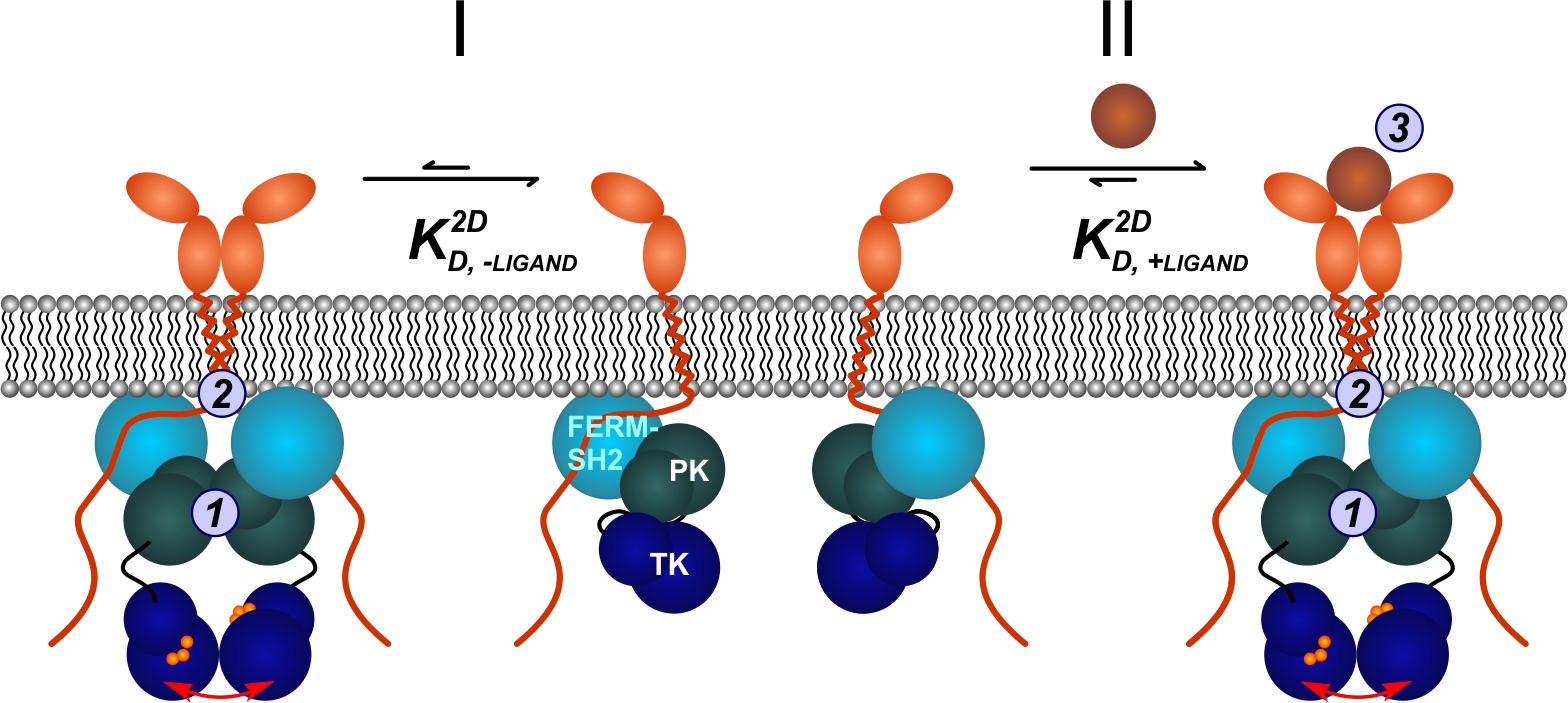
Proposed mechanism of homodimeric cytokine receptor activation deduced from live cell dimerization assays: in the absence of ligand (I), the basal level of dimerization caused by intrinsic weak interactions mediated via the JAK pseudokinase (PK) domains (1) and transmembrane/juxtamembrane domains (2) is negligible at physiological receptor densities. Ligand binding provides the additional binding energy (3) required to shift the equilibrium towards the dimeric state, leading to release of the tyrosine kinase (TK) domain by promoting interactions between the regulatory PK domains (II).
Project P11 - Christian Ungermann
Membrane tethering and fusion at endosome and vacuole
Transport and sorting of proteins and lipids along the endocytic pathway to the lysosome-like vacuole relies on a process called endosomal maturation, where early endosomes change surface composition and morphology before finally fusing with the vacuole. This process requires the coordination of protein complexes responsible for recycling (Retromer), downregulation (ESCRT) and fusion (Rab GTPases, Tethering complexes, SNAREs). These complexes have independent activities, but are likely connected to each other.
Within this project, we will analyze the activation and turn-over of the endosomal Rab5 (in yeast Vps21) and Rab7 GTPases (in yeast Ypt7), the interplay of these Rabs with the CORVET and HOPS tethering complexes, and the crosstalk with retromer and ESCRT complexes on endosomal microcompartments during their maturation. Furthermore, the project will analyze the dynamics of these complexes between membrane and cytosol, the dynamics of their assembly and disassembly on endosomal and vacuolar subcompartments, and their connection to trafficking pathways like the AP-3 pathway. The project takes advantage of yeast as a model organism and several reconstitution approaches to understand the particular function of the involved proteins.
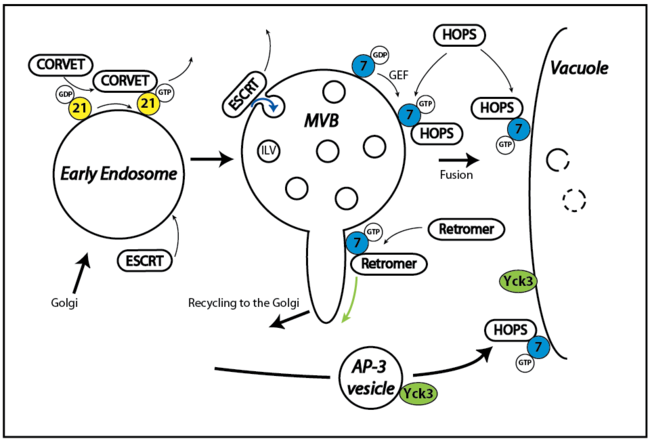
Current model of endosomal maturation and cargo sorting at the endosome - vacuole interface. The early endosome contains the Rab GTPase Vps21 (yellow) and the CORVET tethering complex. Cargo is sorted into intralumenal vesicles by the ESCRT machinery (blue arrows). Retromer binds the late endosomal Rab Ypt7 (blue) and recycles sorting receptors to the Golgi apparatus (green arrows). The HOPS complex facilitates the fusion between MVB and the vacuole. Yck3 (green) reaches the vacuole via the AP-3 pathway.
Project P13 - Sabine Zachgo
Analysis of nuclear ROXY interactions with transcription factors
Glutaredoxins (GRXs) are ubiquitous oxidoreductases implicated in redox homeostasis that catalyze the reversible reduction of disulfide bonds. Until recently, GRXs were known to be mainly involved in oxidative stress responses in plants. Analysis of the Arabidopsis thaliana CC-type GRXs ROXY1 and ROXY2 revealed a novel developmental GRX function, namely regulating petal and anther formation. ROXY1/2 belong to a land plant-specific class of GRXs comprising 21 members with a CC-type active site motif, deviating from the ubiquitously occurring CPYC and CGFS GRX classes. The ROXY1 protein interacts in the nucleus with the TGA transcription factor PERIANTHIA (PAN) and a nuclear ROXY protein activity is indispensable to govern normal flower development. We aim to analyze the spatial and temporal ROXY/PAN interaction dynamics in the nucleus and how they are affected by redox-conditions using different microscopy techniques. The effects of posttranslational protein modifications on ROXY and PAN proteins will be investigated by biochemical studies. In Arabidopsis thaliana, redundant ROXY activities often hinder their functional analyses. The basal land plant Marchantia polymorpha comprises only two ROXY genes. Marchantia is a novel emerging plant model species and will be used together with Arabidopsis in this project to investigate and compare the interactions of ROXY proteins with transcription factors and their nuclear activities during land plant evolution.

The glutaredoxin ROXY1 and TGA transcription factor PAN govern together petal development. Wild type Arabidopsis flowers develop four petals. Lack of ROXY1 activity in the roxy1 mutant leads to a reduced petal initiation, whereas pan mutants form five instead of four petals, indicating opposing effects on promoting petal initiation.
Project P14 - Joost Holthuis
Unravelling the mechanisms and biological impact of sphingolipid homeostasis
Sphingolipids are multipurpose components of cellular membranes with critical roles in cell organization and signaling. This project focuses on the homeostatic mechanisms of sphingolipid metabolism in a microcompartment of the ER and how these mechanisms are integrated in cellular life-death decisions. Following identification of a putative ceramide sensor as ER-resident suppressor of mitochondrial apoptosis, we employed photocrosslinkable lipids to capture novel proteins that participate in ceramide-mediated cell death. Taking advantage of (photo)switchable lipids, lipid carriers and metabolic enzymes, we now seek to: i) elucidate how the newly identified ceramide effector proteins operate; ii) gain optical control over sphingolipid metabolism and signaling in live cells; iii) explore the impact of acute sphingolipid imbalances on cell fate and organization in disease-relevant models.
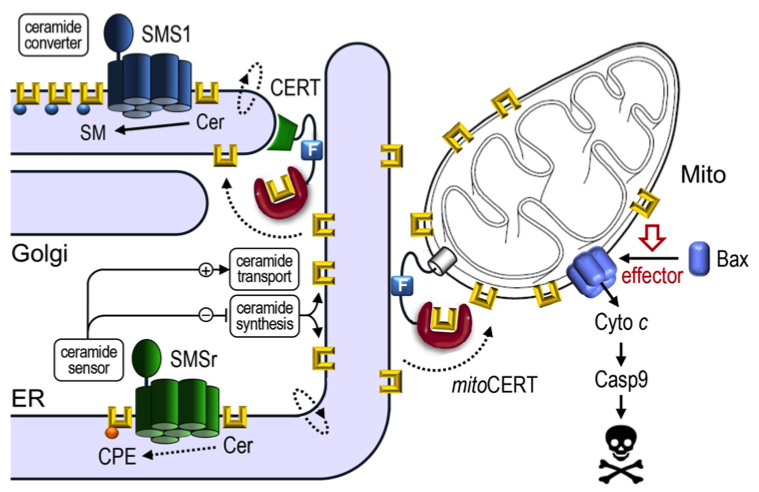
Ceramides synthesized in the ER bypass vesicular trafficking via ceramide transfer protein CERT to reach the Golgi, where they run into a metabolic trap set by sphingomyelin synthase SMS1. SMSr is a SMS1-related protein with dual role as ceramide sensor to keep ER ceramide levels low. Acute inactivation of SMSr triggers Bax-dependent apoptosis due to a rise in ER ceramides and their mislocalization to mitochondria. Expression of a mitochondria-targeted version of CERT, mitoCERT, phenocopies SMSr inactivation, indicating that the arrival of ER ceramides in mitochondria suffices to commit cells to death.
Project P16 - Karin Busch
Impact of mitochondrial ultra-structure and dynamics on OXPHOS functionality and mitochondrial performance: matters of micro-compartmentation
Oxidative phosphorylation (OXPHOS) in mitochondria provides the vast majority of energy for cellular housekeeping, development and differentiation. Its impairment is closely associated with the process of aging, the progress of neuro-degenerative and neuro-muscular diseases and cell death. Our genuine scientific interest is to understand the significance of the spatiotemporal organization and functionality of the oxidative phosphorylation (OXPHOS) system in this context. In particular, we want to decipher the impact of mitochondrial dynamics and ultra-structure on OXPHOS functionality and mitochondrial performance by imaging the relevant processes on the single organelle and single molecule level in human cells.
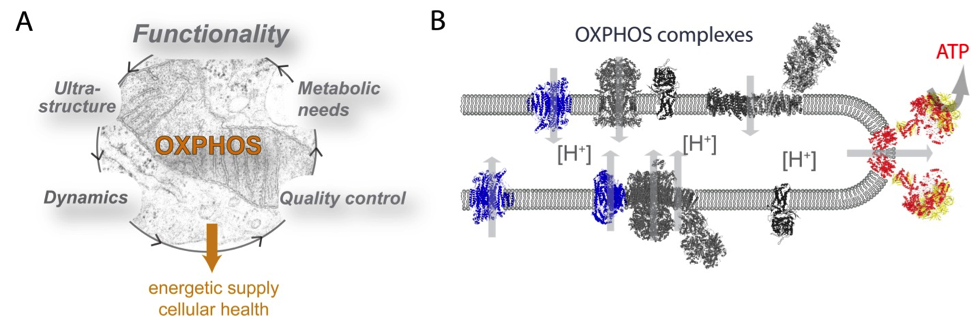
Mitochondrial oxidative phosphorylation (OXPHOS) in the center of bioenergetics. A) OXPHOS functionality is affected by multiple interrelationships: mitochondrial ultrastructure, dynamics, metabolic needs and quality control. B) The current view of OXPHOS complexes in cristae membranes.
Project P17 - Daniel Kümmel
Establishment of endosomal membrane domains by guanine nucleotide exchange factors
Rab GTPases are essential components of the conserved fusion machinery and serve as markers of membrane identity to establish microcompartments in the endomembrane system. Their precise localization is controlled by guanine nucleotide exchange factors (GEFs), which concurrently activate the cognate Rab GTPase and cause its membrane recruitment. Thus, understanding the organization and spatio-temporal coordination of intracellular traffic requires knowledge of the mechanism of RabGEF function. Current models envision that RabGEFs need to integrate several input signals and that a threshold has to be reached to trigger their function. We aim to elucidate how this process is orchestrated at a molecular level for the longin-heterodimer (LH) GEF family, which includes the Mon1/Ccz1 (MC1) and Hps1/Hps4 (BLOC3) complexes. Analysis of complex structure combined with functional studies of protein-protein and protein-lipid interactions will help us to understand how these GEFs define specific subregions in the endosomal system.
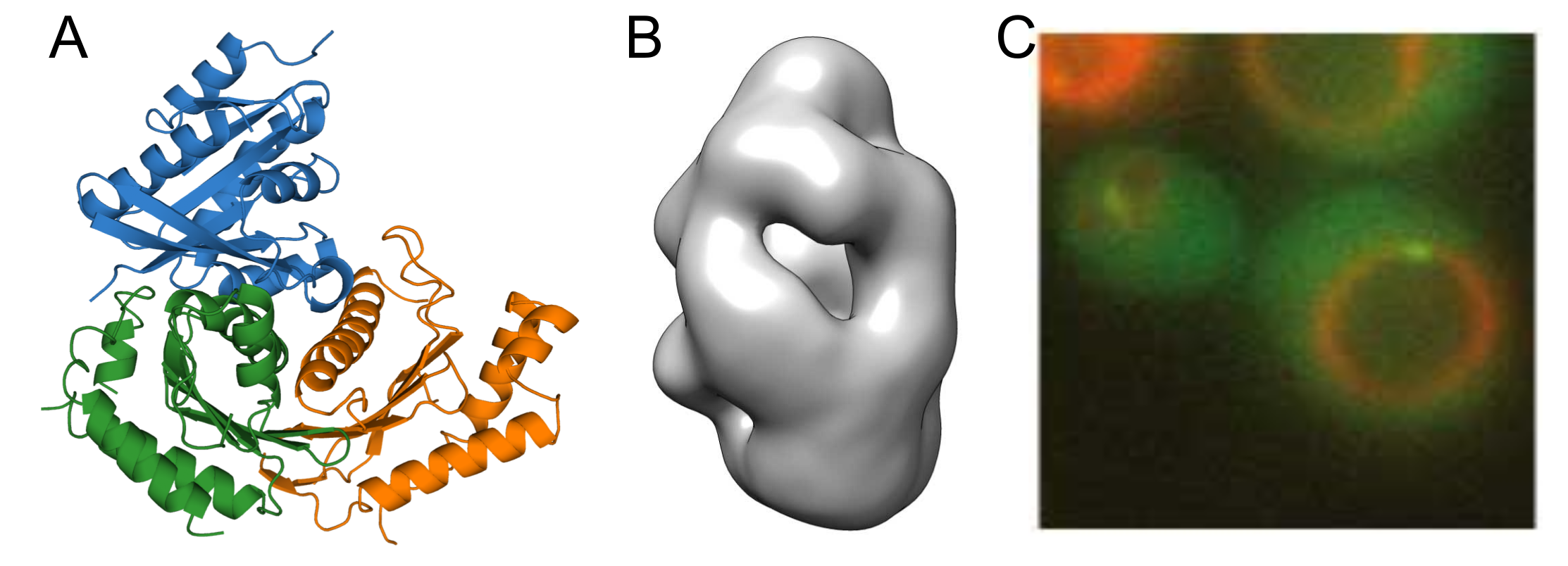
A) Crystal structure of the catalytic Mon1/Ccz1 core complex bound to its substrate Ypt7 B) Negative stain reconstruction of the full-length Mon1/Ccz1 complex. C) Localization of Mon1/Ccz1 to endosomal microcompartments visualized in yeast cells.
Project P18 - Roland Wedlich-Söldner
The role of membrane recycling for the plasma membrane organization - spatial distribution of endocytosis and secretion
Biological membranes have to perform a large variety of functions and are thus organized into a large variety of distinct domains. This project aims to elucidate the role of membrane recycling for the lateral segregation of integral membrane proteins into dynamic microcompartments in the budding yeast plasma membrane (PM).
We have recently used a combination of Total Internal Reflection Fluorescence Microscopy and 2D deconvolution to demonstrate that all proteins in the yeast PM are organized into distinct domains. Importantly, most of the tested proteins either segregated from each other or showed random overlap. However, we found that PM proteins with identical or similar transmembrane (TM) domains co-segregated, and that domain patterns were strongly influenced by cellular lipid composition. We have also shown that a particular domain association can be essential for biological functions of PM-proteins. Our findings indicate that proteins in the yeast PM self-organize into numerous partially overlapping domains. Such a patchwork membrane likely arises from a combination of weak interactions between the diverse lipid and protein constituents of biological membranes.
As all yeast integral PM components that we observed exhibit very slow lateral mobility we expect that delivery and removal of membrane components via exo- and endocytosis have a major influence on the final lateral localization patterns. However, it is so far not at all clear how sites for vesicle fusion and fission at the PM are chosen and to what extent those sites contribute to the formation of PM domains. We will address these issues in two interconnected projects, focussing on endo- and exocytosis, respectively.
Recent publications:
1. Crevenna AH, Arciniega M, Dupont A, Mizuno N, Kowalska K, Lange OF, Wedlich-Soldner R, Lamb DC (2015). Side-binding proteins modulate actin filament dynamics. Elife 4: e04599.
2. Schuberth C, Wedlich-Söldner R (2015). Building a patchwork - The yeast plasma membrane as model to study lateral domain formation. Biochim Biophys Acta 1853: 767-74.
3. Klingner C, Cherian AV, Fels J, Diesinger PM, Aufschnaiter R, Maghelli N, Keil, T., Beck G, Tolic-Nørrelykke IM, Bathe M and Wedlich-Soldner R (2014). Isotropic acto-myosin dynamics promote organization of the apical cell cortex in epithelial cells. J Cell Biol. 207: 107-21.
4. Freisinger T Klünder B, Johnson J, Müller N, Pichler G, Beck G, Costanzo M, Boone C, Cerione RA, Frey E and Wedlich-Soldner R (2013). Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat Commun. 4: 1807.
5. Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Soldner R (2012). Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol. 14: 640-8.

Patchwork organization of the yeast plasma membrane
Project P19 - Christian Kost
Regulation, structure, and function of interbacterial nanotubes
Bacteria frequently engage in contact-dependent interactions, in which cells transfer cytoplasmic materials such as proteins or DNA to other bacterial cells. Recently we discovered that auxotrophic Escherichia coli cells, that were unable to produce certain amino acids, used lipid-based nanotubes to derive cytoplasmic amino acids from other con- or heterospecific cells. This project aims at unravelling the molecular and physiological underpinnings of intercellular nanotubes, particularly focussing on the following main questions: i) Which structural and regulatory genes are involved in the formation of nanotubes?, ii) Which materials are transferred between cells?, iii) How exactly are materials transferred?, and iv) What determines the choice of suitable host cells? These questions will be addressed by combining unbiased ‘omics’- approaches (i.e. genomics, transcriptomics, and proteomics) and a targeted genetic analysis with both superresolution live-cell imaging and electron microscopy of nanotube-forming cells. Together, the expected results will provide fundamental mechanistic insights into the structure and function of the newly discovered interbacterial nanotubes.
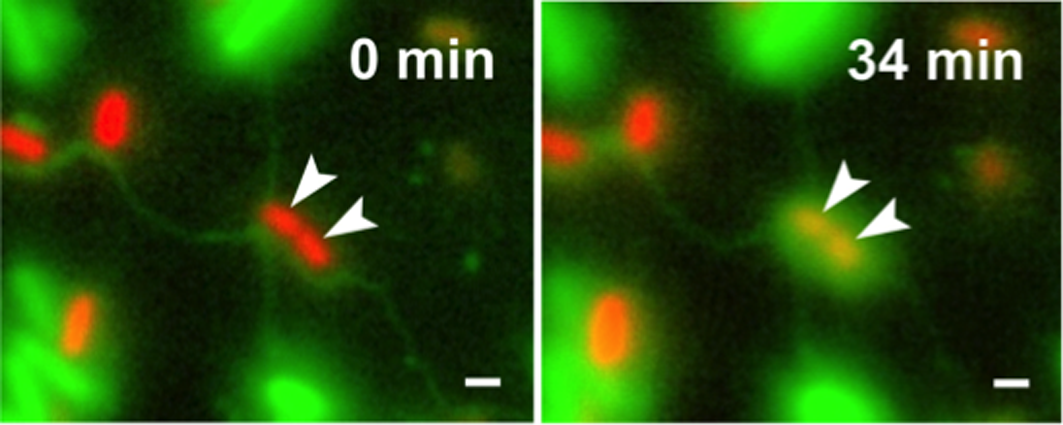
E. coli (green cells) uses intercellular nanotubes to transfer cytoplasmic materials to Acinetobacter baylyi (arrows, red cells) (Pande et al., Nat Commun 2015). Scale bars = 2 µm.
Project P20 – Florian Fröhlich
Mechanisms of sphingolipid export from the endo-lysosomal system
Sphingolipids (SLs) are abundant and essential molecules in eukaryotes that have crucial functions as signaling molecules and as membrane components. They are important for many processes, including endocytosis and surface receptor function. Pathological changes in sphingolipid levels are associated with many common pathologies, ranging from obesity to cancer, asthma, and atherosclerosis, and to neuro-degenerative diseases and genetic disorders. How the cellular distribution of sphingolipids between different organelles is maintained, how this influences sphingolipid homeostasis and how cellular physiology is affected remains unknown. In this project we will combine our expertise in mass spectrometry based proteomics and lipidomics with biochemical, genetic and cell biological assays to shed new light on the regulation of these fascinating lipids in the endo-lysosomal system. We will map the amount of different SL species in yeast vacuoles, we will identify vacuolar SL binding/transport proteins and characterize the mechanism of SL export from the endo-lysosomal system. Together, this will help to understand how cells maintain sphingolipid homeostasis in the endo-lysosomal system.
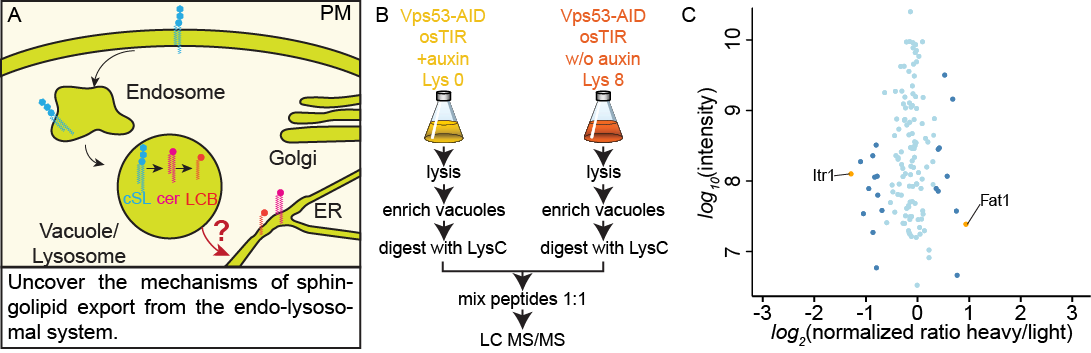
(A) Mechanisms of sphingolipid export from the endo-lysosomal system. (B) Proteomics setup to identify proteins depleted at yeast vacuoles after the induced knockout of Vps53. (C) Vacuolar annotated proteins identified in the proteomics approach. SILAC ratios are plotted against protein intensities. Proteins are color coded according to significance (yellow P<1e-4, steel blue P<0.05, light blue =n.s.)
Project P21 – Heiko Harten
Functional characterization of a novel mechanism regulating SERCA activity and Ca2+ homeostasis in Drosophila melanogaster
Muscle contraction represents a well characterized molecular process that is, to a considerable extent, regulated by variable Ca2+ concentrations within the cytosol. A major player responsible for modulating this concentration is the Sarcoplasmic/Endoplasmic Reticulum Calcium ATPase (SERCA) that transports Ca2+ from the cytosol into the sarcoplasmic reticulum (SR). This activity significantly reduces cytosolic Ca2+ levels and thus marks the beginning of the muscle relaxation phase. Accordingly, tight regulation of SERCA-activity is critical to proper functionality of muscle tissue.
As known for vertebrates and insects, activity of SERCA is regulated by peptides that bind to the enzyme and thereby modulate its activity. Vertebrate and fly loss-of-function mutants for corresponding peptides exhibit impaired Ca2+ transients in heart cells as well as severe heart arrhythmia. Up to now, it is largely unknown how homeostasis and turnover of such peptides are regulated.
We found that increased expression of the peptidase Neprilysin 4 (Nep4) phenocopied the depicted effects on heart physiology. Significantly, Nep4 catalytic activity was essential, thus confirming abnormal peptide hydrolysis as a causative factor. By combining in vitro and in vivo analyses, we identified Nep4 mediated hydrolysis of SERCA regulatory Sarcolamban peptides as the critical physiological event. Furthermore, Nep4 and SERCA coprecipitated and exhibited substantial colocalization with Sarcolamban in membranes of the SR, indicating that close spatial proximity is of high mechanistic relevance.
We currently focus on in-depth analyses of the suborganellar localization and interaction dynamics between Nep4, Sarcolamban, and SERCA. Due to a high evolutionary conservation of involved factors, the project has significant potential to advance the current understanding of muscle physiology and function in health and disease.
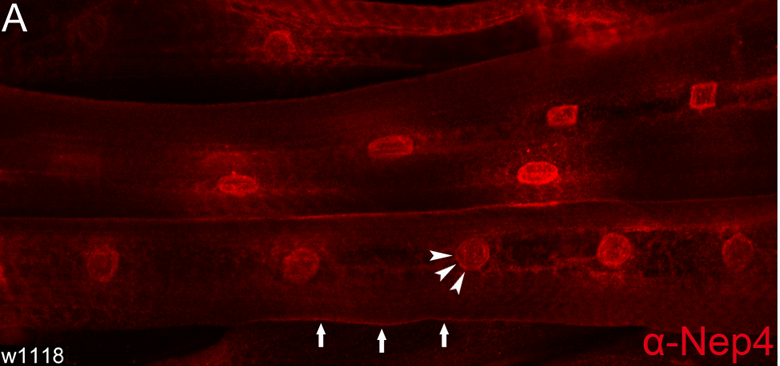
Nep4 localizes to membranes continuous with the nuclear membrane (arrowheads). In addition, a weaker signal at the surface of body wall muscles is apparent (arrows).
STED-based triple labeling of Nep4, SERCA, and Sarcolamban in 3rd instar larval body wall muscles confirms co-localization of all factors in distinct membrane microcompartments of the sarcoplasmic reticulum, particularly around the nucleus (N).
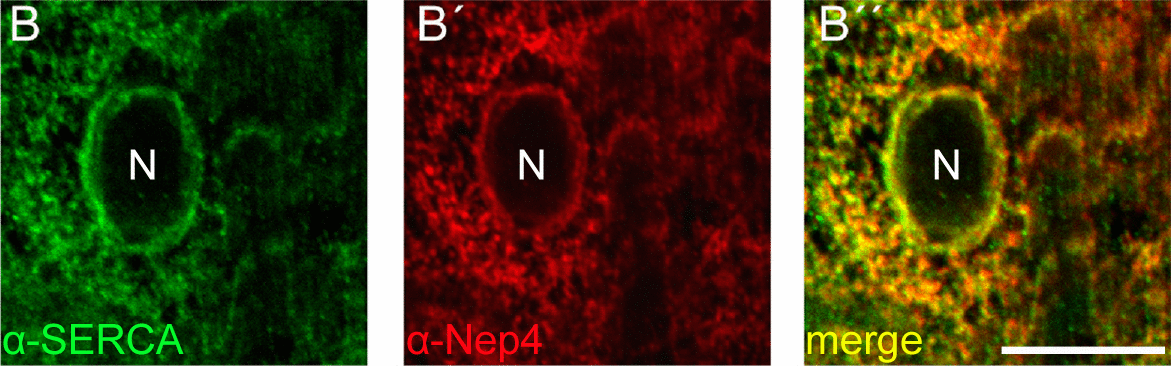
Double labeling with SERCA confirms co-localization of both enzymes in distinct membrane microcompartments of the sarcoplasmic reticulum. N = nucleus. Scale bar = 10 µm.
Project P22 – Milos Galic
Lipid dependence of signaling at curved membrane microcompartments
In this project, we aim to answer how biophysical membrane parameters (e.g. rigidity, micro-viscosity) affect mechano-chemical signaling in living cells (see Figure). Specifically, we will take advantage of a curvature-dependent signaling module that we recently discovered, which mediates continuous leading edge re-initiation and in consequence migration persistence. To determine the membrane contribution to formation and function of this curvature-dependent signaling hub, cellular dynamics and biophysical membrane parameters will be quantified for physiological and pathological membrane conditions. The proposed research is relevant, as it will provide fundamental insights into the interplay between plasma membrane organization and curvature-dependent signaling.
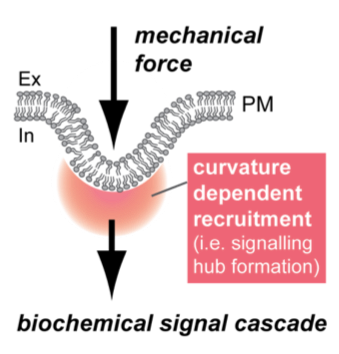
Mechano-chemical signal translation at curved membrane microcompartments. Forces applied from inside or outside of the cell cause plasma membrane deformations. Deformations trigger enrichment of curvature-sensitive proteins and lipids, forming transient signaling hubs at cellular membranes.
Project P24 - Ayelen Gonzalez Montoro
Architecture and function of vacuole membrane contact sites
Membrane contact sites are specialized communication platforms established between organelles, in which the two membranes are closely apposed but do not fuse. Channels and lipid transport proteins localized there enable the exchange of metabolites. Additionally, these structures can influence morphology, movement and positioning of the organelles in the cell.
Microscopy based screens have detected that membrane contact are established between most organelles in the cell. However, many of these structures have not been characterized regarding the proteins that establish them or their function. We performed a microscopy-based screen to identify components of the vacuole-mitochondria membrane contact site in the yeast Saccharomyces cerevisiae. We found an uncharacterized protein, which localizes at this interface. Further work showed that this protein also localizes to the vacuole-peroxisome contact site and to the vacuole-endoplasmic reticulum contact site, so we have named it Cvm1 (contacts of the vacuole membrane 1).
Within this project, we will address how Cvm1 establishes these different contact sites, which proteins it interacts with on the different organelles, and which domains within the protein are required for these interactions. We will also focus on the function of Cvm1 and the contact sites it forms. We will determine its role in contact site and organelle turnover during growth and stress, and its relations with other cellular processes by looking at the consequences of its deletion and its genetic interactions. This project will thus provide novel insights on how distinct vacuole-organelle contact sites affect the functionality of the involved organelles and thus cell physiology.
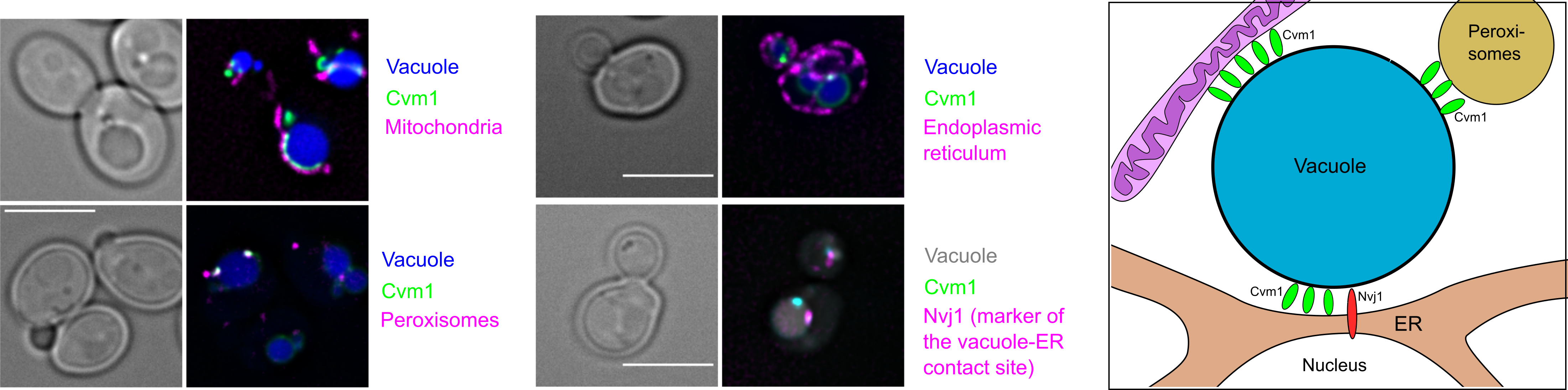
Fluorescence microscopy of live Saccharomyces cerevisiae cells shows the localization of Cvm1 in the interface between vacuoles and mitochondria; vacuoles and peroxisomes; and vacuoles and the endoplasmic reticulum. Additionally, co-localization of Cvm1 with the known marker of the endoplasmic reticulum-vacuole contact site Nvj1 is shown. The cartoon depicts the localization of Cvm1 to these 3 different contact sites of the same organelle.
Project 25 - Caroline Barisch
Lipid Trafficking at Membrane Contact Sides during Mycobacteria Infection
Tuberculosis is caused by Mycobacterium tuberculosis (Mtb) and is responsible for 1.6 million deaths worldwide every year. The high lipid content of this pathogen accounts for many of its unique clinical manifestations. One of the main characteristics of Tb is the formation of lipid-loaded, foamy macrophages during chronic infection. A growing body of evidence indicates that Mtb mobilizes lipid droplets (LDs) to scavenge lipids from its host cell. However, how this pathogen remodels the lipid metabolic network of the host to support its persistent lifestyle is so far poorly understood.
An emerging concept is that bulk lipid transport inside eukaryotic cells largely occurs at membrane contact sites (MCSs), specialized microcompartments where two organelles are closely apposed to facilitate a functional integration of compartmentalized cellular processes. MCSs are typically enriched in lipid biosynthetic enzymes and transport machinery, notably cytosolic lipid transfer proteins (LTPs) that enable lipids to reach their destination independent of vesicular trafficking. Intriguingly, pathogens like tombusviruses and Chlamydia have been reported to co-opt LTPs from their hosts to build MCSs between the inclusion membrane and host organelles for the acquisition of lipids needed for their replication.
Within this project we will use advanced microscopy techniques and lipidomics to unravel the role of pathogen-induced MCSs during M. marinum infection of Dictyostelium. In addition, we will take advantage of clickable and fluorescent lipid probes to monitor lipid transfer from the host to the intracellular pathogen via MCSs.
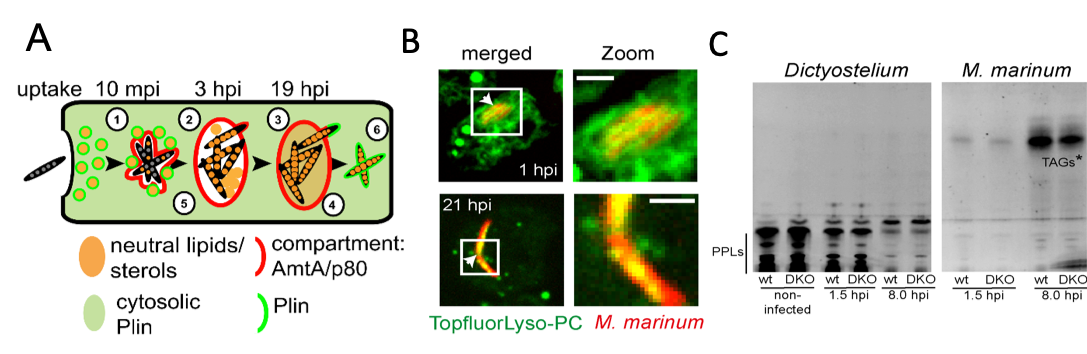
A) Schematic representation of neutral lipids during M. marinum infection of Dictyostelium. LDs translocate to the MCV (1) leading to the enrichment of LD-like structures (2) and neutral lipids (3) inside the MCV. Bacteria accumulate LDs during infection (4). Bacterial lipids are at least partially derived from the host (5). Perilipin (Plin) labels cytosolic bacteria as soon as the bacteria are in direct contact with the cytosol (6). B) Topfluor-LysoPC-tagged host lipids first label the membrane of the MCV and are subsequently found inside the bacteria C) TLCs showing that M. marinum incorporates fluorescently-labelled fatty acids released from host phospholipids (PPLs) into triacylglycerols (TAGs*). Images from: Barisch et al. (2015), Barisch and Soldati (2017).
Project P26 - Katia Cosentino
Molecular mechanisms of pore formation by Gasdermin D in pyroptosis
Pyroptosis is a highly inflammatory form of regulated cell death initiated in response to detection of pathogens or endogenous danger signals in the cell. Gasdermin D (GSDMD) and other members of the Gasdermin family execute pyroptosis by forming membrane pores that permeabilize the plasma membrane (PM). This step in pyroptosis is central to mediate the secretion of inflammatory cytokines into the extracellular space to elicit an immune response, and eventually lead to cell death, thus exposing pathogens to effector cells of the immune system.
The goal of this project is to elucidate the molecular mechanisms of GSDMD membrane pore formation during pyroptosis. We aim: 1) to identify and characterize PM constituents responsible for GSDMD membrane recruitment and function; 2) to dissect the steps of GSDMD assembly into oligomers and to provide a structural characterization of GSDMD pores; 3) to correlate protein stoichiometry with pore functionality to understand pyroptosis in detail. Together, these studies will broaden our knowledge on the role of GSDMD in modulating inflammatory cell death.
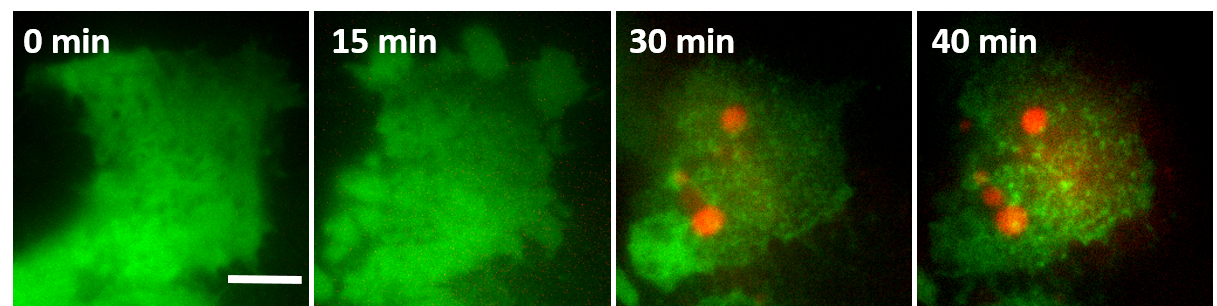
GSDMD at the plasma membrane (PM) of a pyroptotic cell. Initially, GSDMD (green) is homogenously distributed in the cytosol of the cell. Pyroptosis induction (at time 0) induces its translocation and assembly (green dots) at the PM. GSDMD oligomerization is concomitant with PM permeabilization (indicated by DNA staining by the membrane impermeable dye PI, red). Scale bar: 5 μm.
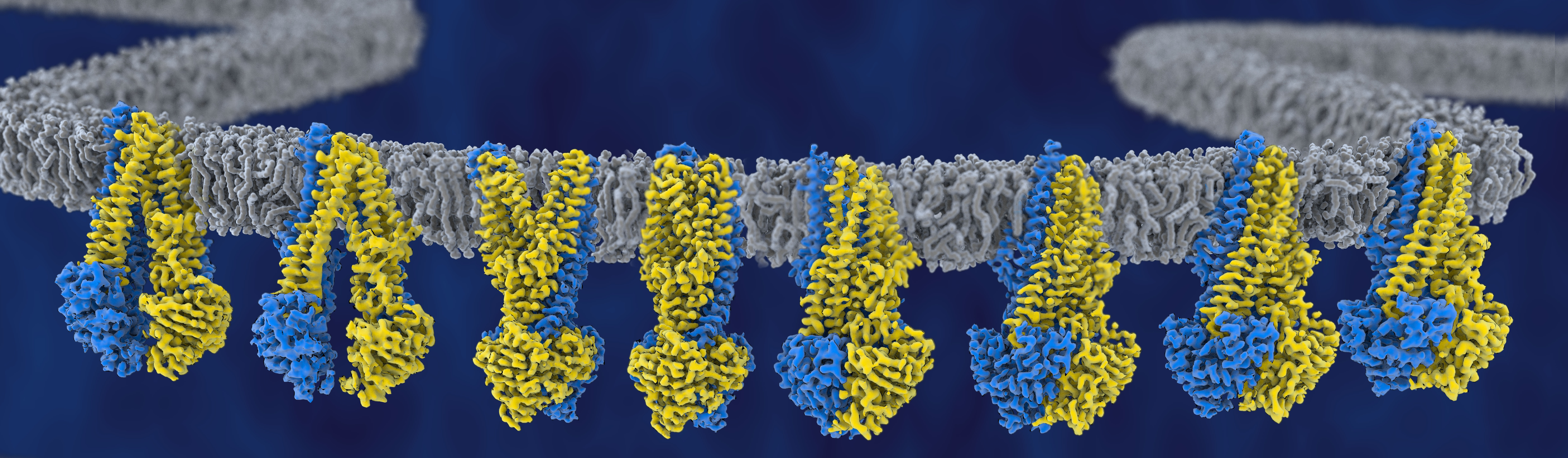
Project 27 - Arne Möller
ABC exporter function within their lipid microenvironment
ATP binding cassette (ABC) exporters are essential contributors to the emergence of multidrugresistance. They form a microcompartment with their surrounding lipids, and the composition of this microenvironment directly affects their conformational spectrum, as well as their transport efficiency. The molecular causes of this remain unknown, as thorough comparative studies, recapitulating the protein-lipid composition of the microcompartment are challenging. Using cryo-EM, we have recently described the conformational spectrum of the heterodimeric ABC transporter TmrAB in a lipid environment and were able to address multiple long-standing controversies. With this project, we will systematically characterize the impact of microcompartment composition on the dynamics of a homodimeric ABC exporter. We will investigate how the transporter initiates binding to a substrate and structurally determine its conformations in different hydrophobic environments. We expect that our research will have far-reaching implications in ABC transporter biology and of conformational dynamics of membrane proteins in general. This project will add a high-resolution angle to the description of microcompartments and will contribute the method of cryo-EM to the research center.
Project INF - Michael Hensel
Information Infrastructure for SFB 944
The majority of projects in SFB 944 make use of state-of-the-art multimodal imaging techniques, leading to the generation of large volumes of raw and processed experimental data. For the accessibility of these data for project participants and the scientific community, safe storage and long term archiving in compliance with DFG requirements, the existing storage and archive infrastructure will be extended. We propose to adapt the open source database suite OMERO to the local environment of the SFB 944, CALMOS and the research building CellNanOs that will be in operation in 2016. Image files and metadata from the various microscopy systems used for SFB 944 projects will be handled via the OMERO database. For storage of the increasing volume of experimental data, a network attached storage (NAS) is requested that will be operated via the OMERO interface. For backup and archiving, a tape library system is requested. This large volume long-term storage system is operated by the Rechenzentrum of the University Osnabrück (RZ) and is compatible with existing storage solutions of the RZ. Implementation of the OMERO database allows use of a single platform for data storage and archiving, metadata handling and access from internal and external users. The INF project will provide data archival and access function for compliance to DFG regulations on availability of data generated in DFG-funded projects.
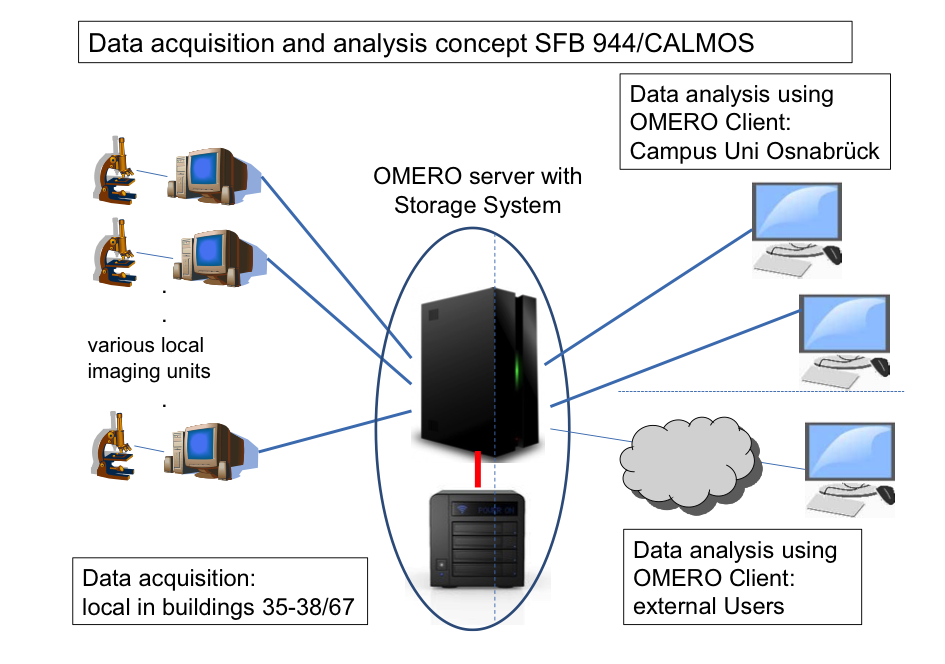
Data storage and archiving concept for INF. Various local imaging systems acquire image data that are locally stored during acquisition sessions. Via the high-speed network, data are immediately moved to a NAS server operated with an OMERO platform adapted to the SFB 944/CALMOS environment. At that stage, data are subject to automated backup routines. Depending on the form of data and access frequency, data are moved from the storage server to archives in the tape library. The scalable design will allow integration of further imaging systems as well as an extension of NAS and tape library storage. Network bandwidth is indicated by lines (blue 1 Gbit; red, 10 Gbit).
Advanced imaging techniques for resolving cellular microcompartments. (a, b) Single molecule localization microscopy. (a) Individual emitters can be localized beyond the diffraction limit with a typical precision of 20-30 nm. (b) Based on this concept, superresolved images can generated by using switchable fluorophores (PALM, STORM). By tracking a limited ensemble of diffusible molecules over time, the spatiotemporal dynamics of molecules and the connectivity within microcompartments can be obtained in addition to superresolved (tracking and localization microscopy, TALM).
Project Z - Karin Busch, Michael Hensel, Jürgen Klingauf, Stefan Kunis, Jacob Piehler
Advanced imaging techniques
Exploring cellular microcompartments fundamentally requires imaging techniques capable to resolve nanoscopic structures. Within the Z-project we aim to provide access to advanced fluorescence and electron microscopy. In the first funding period, we have built up a technological and methodological infrastructure for single molecule localization-based super-resolution (SR) imaging techniques. In the current funding period, we aim to further optimize and apply these existing tools within the consortium. We will in particular focus on three-dimenisonal SR imaging based on a Bessel beam single plane illumination microscope. To increase the temporal resolution, we plan to extend deconvolution based on compressed sensing for three-dimensional single molecule localization. Advanced fluorescence microscopy will be complemented by electron microscopy approaches for ultrastructural characterization of cellular microcompartments. Here, we plan to provide three-dimensional imaging by transmission electron tomography and scanning electron microscopy with sectioning by a focussed ion beam. We will establish cryofixation and freeze substitution techniques and we will develop and apply approaches for correlative light and electron microscopy.
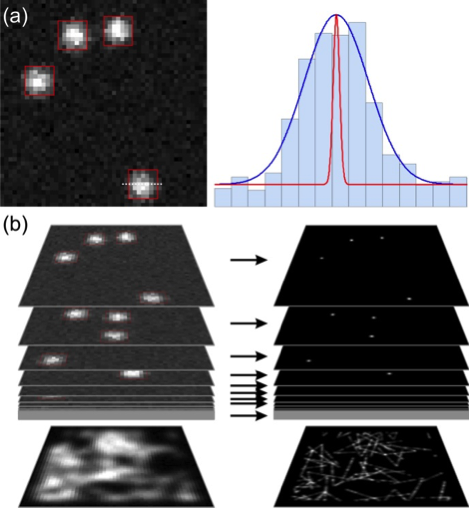
Advanced imaging techniques for resolving cellular microcompartments. (a, b) Single molecule localization microscopy. (a) Individual emitters can be localized beyond the diffraction limit with a typical precision of 20-30 nm. (b) Based on this concept, superresolved images can generated by using switchable fluorophores (PALM, STORM). By tracking a limited ensemble of diffusible molecules over time, the spatiotemporal dynamics of molecules and the connectivity within microcompartments can be obtained in addition to superresolved (tracking and localization microscopy, TALM).
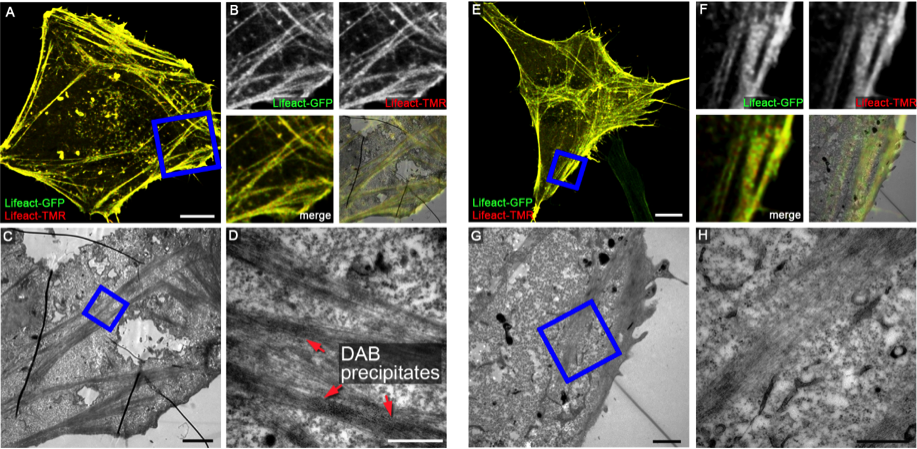
Examples of HaloTag-mediated DAB photoconversion. HeLa cells transiently expressing LifeAct-GFP (green) and LifeAct-HaloTag were fixed and stained with TMR-labelled HaloTag substrate (red). Samples shown in A-D were subjected to DAB photoconversion, samples shown in E-H are controls without DAB photoconversion. Higher magnifications of ultrathin sections shown in C and G indicate the presence of electron-dense DAB precipitates adjacent to F-actin filaments. Scale bars: 10 µm (A, E), 2 µm (C, G), 500 nm (D, H).
Project IRTG - Achim Paululat, Joost Holthuis, Roland Brandt, Christian Kost, Sabine Zachgo
Integrated research training group Physiology and Dynamics of Cellular Microcompartments
The Integrated Research Training Group (IRTG) of the Sonderforschungsbereich (SFB; "Collaborative Research Center") will provide a high-standard and structured interdisciplinary education and training program for doctoral students within the dedicated topic of “Physiology and Dynamics of Cellular Microcompartmentsâ€. The training group will be an active part of the Graduate School of the University of Osnabrueck (ZePrOS). Doctoral students will obtain a broad training in physical, biophysical, biochemical and cell biological techniques as well as in mathematical image and data analysis. Internationalization will be fostered by a student exchange program with partners involved in new as well as already established collaborations.
Former Projects (2011 - 2017)
Project P2 - Guy Hanke
Mechanism of Ferredoxin Reductase - Membrane Interaction, and its Effect on Photosynthetic Partitioning
Efficient photosynthesis depends upon the regulated assembly and disassembly of micro- compartments, composed of super-complexes and electron carriers, at the thylakoid membrane. Super-complexes, able to specifically perform linear or cyclic photosynthetic electron flow have been identified, and all contain the enzyme ferredoxin NADP+ reductase (FNR), which is also present as a soluble protein. In this project, we will analyze the dynamics of FNR association with different complexes, and investigate the structural mechanism that regulates FNR recruit- ment and release from FNR binding proteins. In addition, the impact of these processes on photosynthesis will be examined.
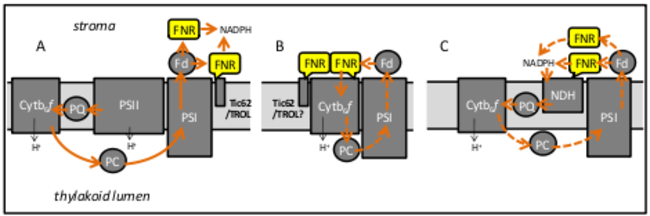
Localization and function of FNR in different PET micro-compartments. A, Micro- compartment for linear PET (solid arrows): electrons flow from photosystem II (PSII), through plastoqui- none (PQ) and the cytochrome b6f (Cytb6f) complex to PSI, where they are donated to Fd and used by FNR to reduce NADP+. FNR that is soluble, associated with PSI, or tethered by the FNR binding proteins Tic62 and TROL could mediate this. B and C Micro-compartments for cyclic PET (hashed arrows): electrons are returned to the PET chain, either directly by Fd (B), or by NADPH, via the NAD(P)H dehydrogenase complex (NDH) (C) generating a proton gradient with no net reductant.
Project P3 - Jürgen Heinisch
Cell wall synthesis and cytokinesis in yeast - the role of sensors and cytokinesis regulators in the microcompartment of the bud neck
Yeast cytokinesis occurs at the microcompartment of the bud neck and involves the controlled synthesis of cell wall components. How the vital processes of cell cycle control (with the final event of cytokinesis) and signalling for cell wall biosynthesis are exactly coordinated remains to be determined. For this purpose, we intend to build on our previous research in both areas and the expertise in yeast genetics. The project will first focus on the sensors of the cell wall integrity (CWI) signal transduction pathway and their relation to the control of the spatial and temporal organization of protein complexes and regulatory components during yeast cytokinesis. A number of imaging techniques will be applied, like fluorescence microscopy and single-molecule AFM. Genetic screens, epistasis analyses and site-directed mutageneses with the main CWI sensors (Wsc1, Wsc2 and Mid2) and the cytokinesis regulator Inn1 will be used to identify new components involved in the regulatory networks in vivo.
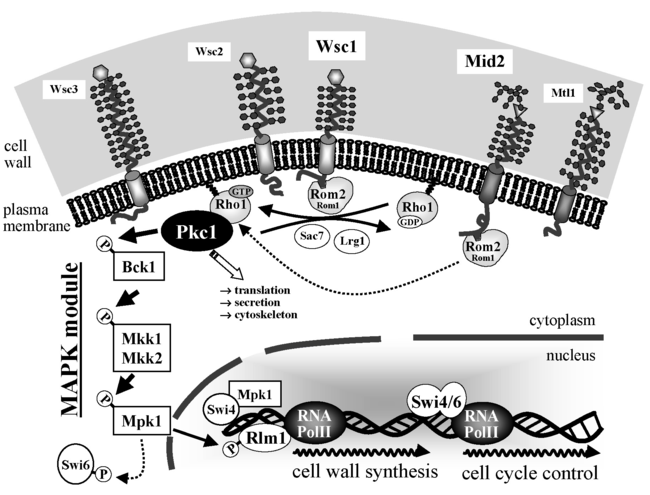
Cell wall integrity signalling in the yeast S. cerevisiae. The CWI sensors (Wsc1- Wsc3 on one hand, and Mid2 and Mtl1 on the other), interact with the GDP/GTP exchange factor Rom2 for the small GTPase Rho1. The latter interacts with the sole yeast protein kinase C (Pkc1), which activates a subsequent MAPK module by phosphorylation of its upper MAPKKK Bck1. Finally, phosphorylated Rlm1 governs the transcription of some 30 genes, whose products are relevant for cell wall construction. Cell cycle pro-gression is controlled by the SBF transcription factor, a heterodimer formed by Swi4/Swi6 (from: Rodicio and Heinisch, 2010).
Project P6 - Hans Merzendorfer
Analysis of the CaaX processing machinery from yeast
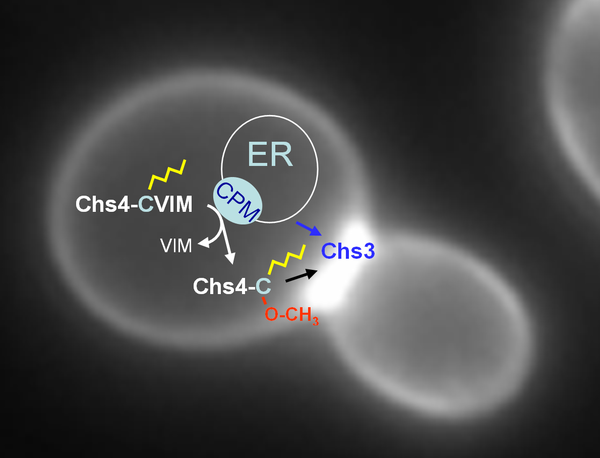
CaaX processing is a posttranslational modification of the carboxyl-terminus of more than one hundred proteins in eukaryotes that are essential for growth, development and survival. It involves isoprenylation of the CaaX motif's cysteine, proteolytic release of the aaX peptide and carboxyl-methylation of the resulting isoprenylcysteine. Focus of this project is the CaaX processing microcompartment transiently formed at the ER, which is required for the modification of yeast Chs4, a regulatory subunit of the chitin synthase (CHS) III complex. The CHS III complex synthesizes chitin for the cell wall and the chitin ring produced at polarized growth sites. We analyze the protein interactions between the constituents of this microcompartment in vitro, and the dynamics of complex assembly and disassembly in vivo. By addressing CaaX processing of Chs4 we try to increase our current understanding on maturation and trafficking of proteins involved in chitin biosynthesis, but also aim to shed light on the still enigmatic biological function of the post-prenylation steps of CaaX processing.
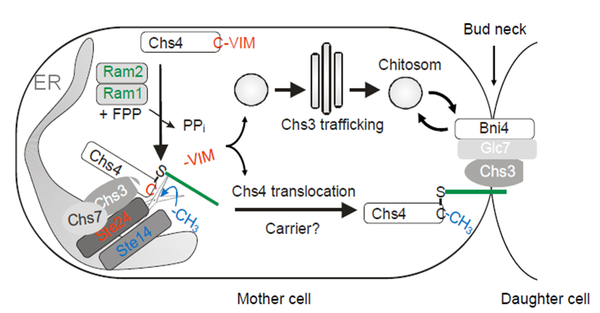
Current model of CaaX processing of Chs4 at the ER. Chs4 is prenylated by Ram1/Ram2 in the cytosol leading to its association with the membranes of the ER. Chs3 recruits Chs4 to Ste24, which cleaves off the aaX peptide allowing carboxy-methylation of the isoprenoid cysteine by Ste14.
Project P9 - Renate Scheibe
Transfer of redox-signals from chloroplast to nucleus by transient microcompartmentation of metabolic enzymes
Cytosolic NAD-glyceraldehyde 3-P dehydrogenase (NAD-GAPDH, GapC) in Arabidopsis thaliana is redox-sensitive and reversibly inactivated by S-glutathionylation and/or S-nitrosyla- tion. In addition to its role in glycolysis as soluble enzyme, GapC can bind to cytoskeleton, mitochondria, and nuclear substructures. We propose it to be involved in retrograde signaling upon increased electron pressure in high light and subsequent oxidative stress, inducing increased expression of nuclear encoded chloroplast NADP-malate dehydrogenase. In this project we plan (i) to correlate the redox-states of the pertinent compartments with the subcellu- lar localization of GapC (as a fusion with GFP) and its varying functions, (ii) to characterize the spatio-temporal dynamics of the cytosolic and nuclear GapC-containing microcompartments, and (iii) to finally determine their special functions in redox signaling and gene expression.
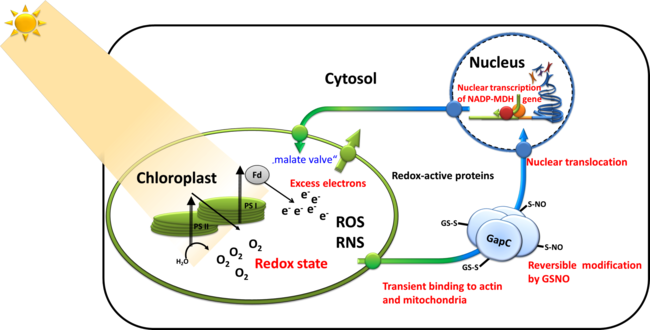
Retrograde signaling of chloroplast redox-state to nucleus. Working model for the stress-induced expression of nuclear encoded NADP-MDH: Excess electron pressure in the chloroplasts leads to ROS/RNS formation, generating nitroso-glutathione (GSNO) that readily modifies sensitive thiols such as those of GapC. This reaction and its reversal are most likely catalyzed by "redoxins". Changed properties of the modified cytosolic enzymes is supposed lead to altered microcompartmentation, and possibly to new functions, namely in gene expression, e.g. of NADP-MDH as part of the malate valve relieving electron pressure in the chloroplasts. Its increased expression will be used as a read-out for the investigated redox-signaling cascade. In this proposal, we therefore ask the question: What is the sequence of events in vivo and what are the particular functions of GapC in this process?
Project P10 - Heinz-Jürgen Steinhoff, Johann Klare
Conformational dynamics of signal transducers in photo- and chemotaxis
Intra- and intermolecular signal transduction mechanisms in membrane-embedded (photo-) receptor/transducer complexes are studied using the photoreceptor/transducer complex NpSRII/NpHtrII, which is responsible for the photoactive behavior of the halophilic archaeon Natronomonas pharaonis. The structure of NpSRII/NpHtrII is related to bacterial chemoreceptors, which were shown to form microcompartments in the cell membranes at the poles of the cell. Concomitantly, the NpSRII/NpHtrII complex will serve as model system for development of new site-directed spin labeling EPR methods to elucidate the structure and conformational dynamics of membrane proteins and protein clusters in vitro and in vivo.
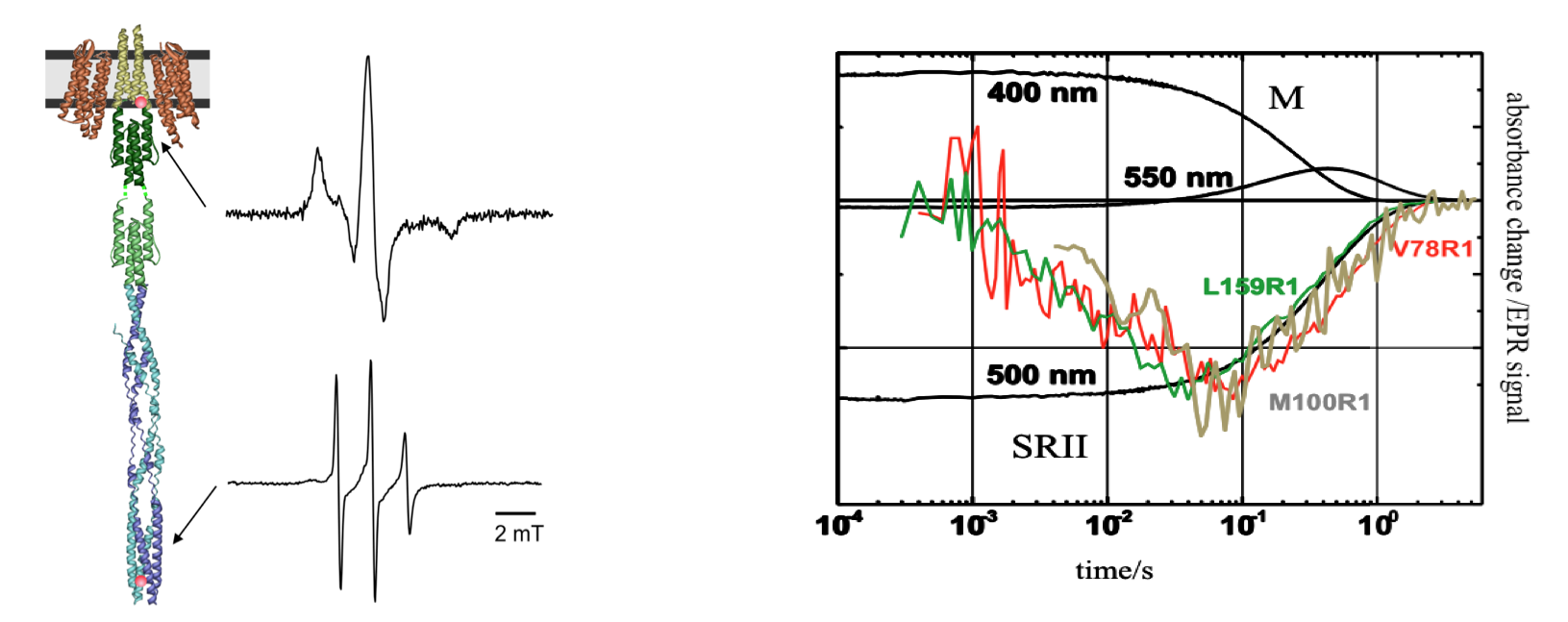
Left: EPR spectra of NpSRII/NpHtrII reconstituted into nano-lipoprotein particles with spin labels bound to position 78 at the water-lipid interface of TM2 (top), and to position 351 in the signaling tip of the transducer (bottom). The EPR spectrum of the spin label at position 351 reveals the high dynamics of the signaling tip. Right: Light induced EPR signal changes for spin labels bound to positions 159NpSRII (green), 78NpHtrII (red) and 100NpHtrII show the transfer of the signal from the receptor NpSRII to the transducer NpHtrII occurring during the M photocycle intermediate monitored at 400 nm.
Its versatile usage has established channelrhodopsin-2 (ChR2) as the most prominent optogenetic tool. The structure and photocycle of this light activated cation channel are related to those of NpSRII. Interspin distance measurements revealed a light induced movement of helix B, which is not observed for other rhodopsins, but is essential for the opening of the ion pore. Time resolved measurements of the transient conformational changes will unravel their relation to the photocycle and provide information about ChR2’s functional mechanism.
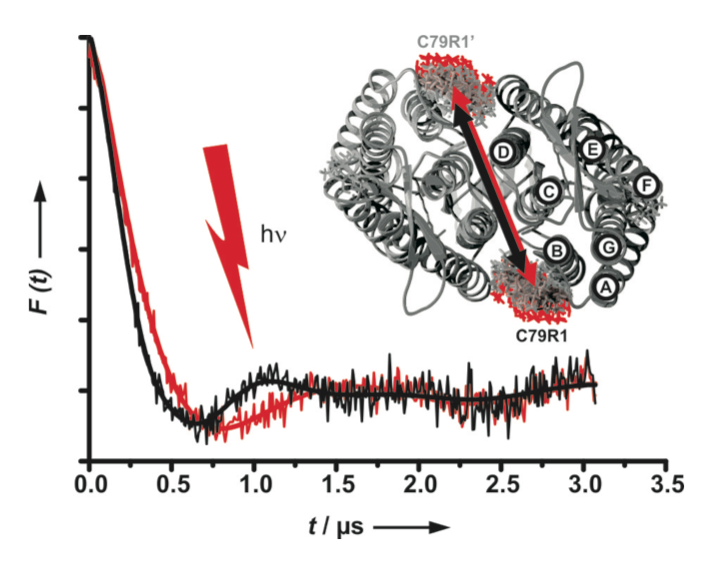
DEER experiments in both the dark-adapted state (black) and upon light activation (red) reveal a change in the distance between helices B and B’ of the ChR2 dimer.
Recent related publications
- Klare JP, Bordignon E, Engelhard M, Steinhoff HJ (2011) Transmembrane signal transduction in archaeal phototaxis: The sensory rhodopsin II-transducer complex studied by electron paramagnetic resonance spectroscopy. Eur J Cell Biol 90:731-739.
- Klose D, Klare JP, Grohmann D, Kay CWM, Werner F, Steinhoff, HJ. (2012) Simulation vs. Reality: A Comparison of In Silico Distance Predictions with DEER and FRET Measurements. PLoS ONE 7(6): e39492. doi:10.1371/journal.pone.0039492
- Sattig T, Rickert C, Bamberg E, Steinhoff HJ, Bamann C. (2013) Light-induced Movement of the Transmembrane Helix B in Channelrhodopsin-2. Angewandte Chemie Int. Edition 52:9705-8.
- Klose D, Voskoboynikova N, Orban-Glass I, Rickert C, Engelhard M, Klare JP, Steinhoff H-J (2014) Light-induced switching of HAMP domain conformation and dynamics revealed by time-resolved EPR spectroscopy. FEBS Letters 588: 3970-3976.
- Orban-Glaß I, Voskoboynikova N, Busch KB, Klose D, Rickert C, Mosslehy W, Roder F, Wilkens V, Piehler J, Engelhard M, Steinhoff H-J, Klare JP (2014) Clustering and dynamics of phototransducer signaling domains revealed by SDSL EPR on SRII/HtrII in membranes and nanodiscs. Biochemistry 54: 349-362.
Project P12 - Helmut Wieczorek
Regulation of V-ATPases by reversible dissociation
Vacuolar H+-ATPases (V-ATPases) are a constituent part of every eukaryotic cell. They are heteromultimeric proton pumps, occur in many different membranes and are essential for a multitude of different biological functions. V-ATPases exhibit a unique type of regulation, the reversible disassembly of the ATP hydrolyzing V1 complexes from the membrane bound, proton translocating VO complexes. Proper transfer of V1 complexes or subunits from the membrane to the cytoplasm and back appears to require not only phosphorylation events, conformational changes in the V1VO holoenzyme, the V1 complex and the V1 subunit C, but also the temporary interaction with protein kinase A, cytoskeletal elements, the heterotrimeric protein complex RAVE and the glycolytic enzyme aldolase. The project focuses on the dynamics of transient microcompartments formed by the V-ATPase holoenzyme, V-ATPase subcomplexes and subunits together with the mentioned proteins, with the aim to understand the mechanisms leading to this unique type of physiologically relevant regulation. For this purpose analyze, on the one hand, the interaction of the V-ATPase, its subcomplexes and subunits with the mentioned proteins biochemically and by cryo-electron microscopy. On the other hand, we focus on in vivo/in situ studies using the yeast V-ATPase in the vacuolar membrane as well as the insect plasma membrane V-ATPase. We choose this comparative approach because in contrast to yeast, where the active, assembled holoenzyme in the vacuolar membrane is the default state, the insect plasma membrane V-ATPase is normally inactive and is activated by hormone-induced reassembly.
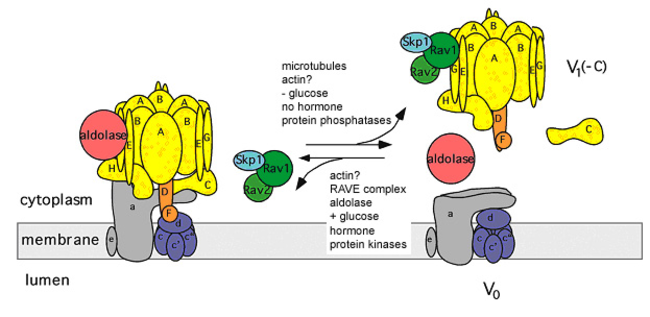
Regulation of V-ATPase activity by reversible disassembly. Modified according to Toei et al. (2010).
Project P15 (old) - Sabine Hunke
Coordination of bacterial two component system signalling
Two-component systems (TCSs) play a pivotal role for bacteria in stress regulation and adaptation, but how these systems are modulated to meet bacterial demands is not well understood. Especially, the role of accessory proteins within the coordination of TCS response is only partly understood.
We investigate the E. coli Cpx-TCS as a model for a microcompartment within and between bacterial membranes. The Cpx-TCS, consisting of the sensor kinase CpxA and the response regulator CpxR, responds to perturbation of the bacterial envelope. Two accessory proteins orchestrate the Cpx-TCS: The periplasmic protein CpxP inhibits the Cpx-TCS in unstressed cells and detects misfolded envelope proteins; the outer membrane lipoprotein NlpE activates the Cpx-TCS during biofilm formation. We demonstrated direct interaction between CpxP and CpxA in unstressed cells and showed that this interaction is mitigated by high NaCl concentration and misfolded envelope proteins (Tschauner et al., 2014).
Now, we will investigate how the opposite functions of CpxP and NlpE are coordinated. We will continue to characterize how CpxP and CpxA communicate. In addition, to understand how communication between two membranes is mediated by the Cpx-TCS, we will analyze the interaction between NlpE and CpxA. Finally, to understand the coordination of signalling, we will study the plasticity of accessory protein dependent and independent signals for the Cpx-reponse.
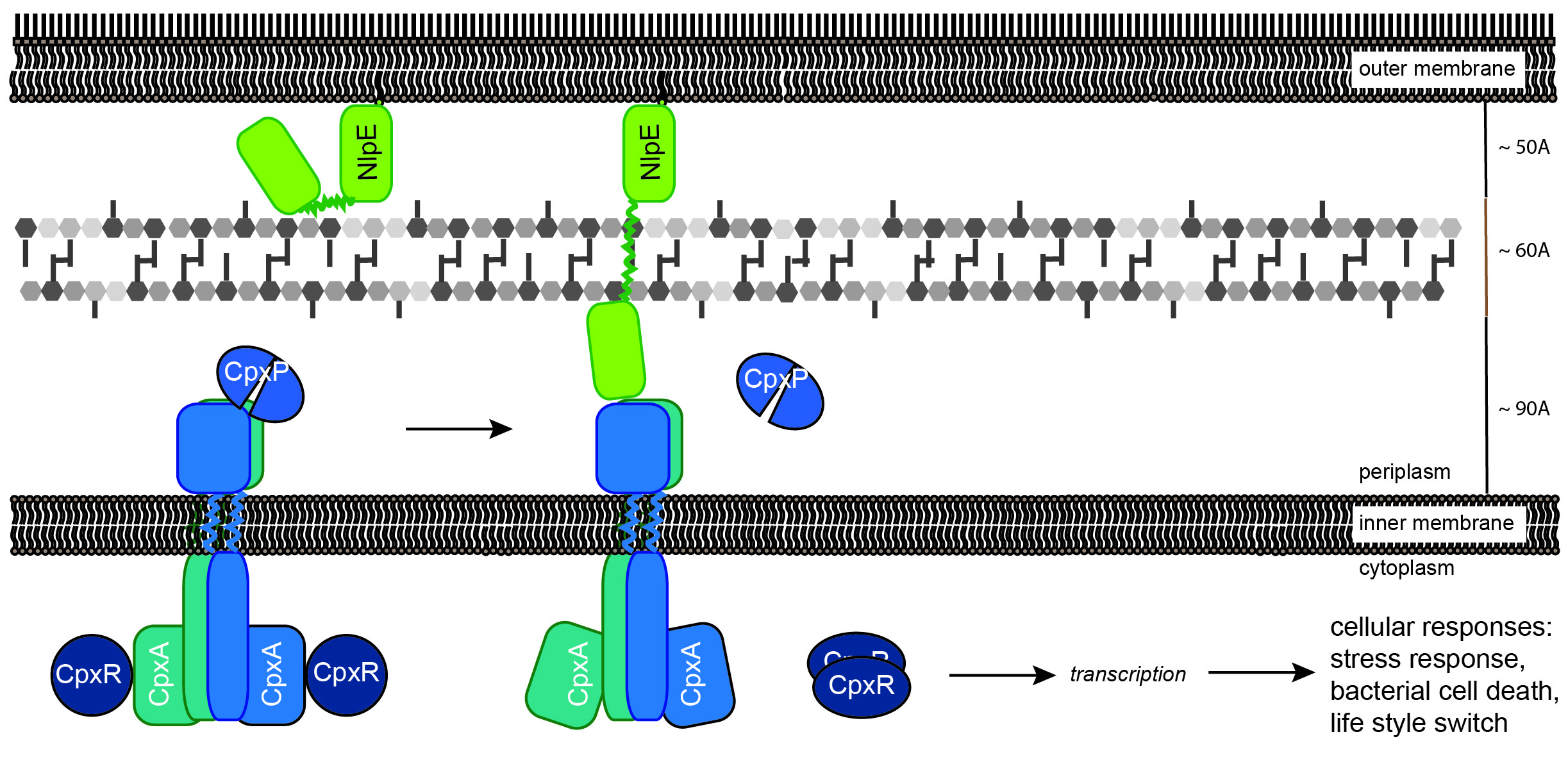
Model depicting proposed functions of the accessory proteins CpxP and NlpE for stimulus perception by the Cpx system Direct interaction between a CpxP dimer and CpxA keeps the sensor kinase in an “Off” mode. Envelope stress such as a high salt concentration or misfolded surface proteins induce the release of CpxP from CpxA resulting in activation of the Cpx-system. The outer membrane lipoprotein NlpE monitors attachment of the bacterial envelope to surfaces. It is speculated that Cpx-system activation by NlpE might result from tertiary or quaternary structural instability causing a direct contact between the C-terminal domain of NlpE and CpxA.